About DeepTrees 🌳
The efficient management of trees relies on precise and current information, encompassing a detailed account of individual crown canopy cover (CC). Canopy cover is a fundamental metric that characterises the horizontal extent and arrangement of tree crowns, commonly expressed as the percentage of total ground area covered by a two-dimensional projection of the tree canopy. Due to its ease of determination and conceptual simplicity, CC is a prominent objective in various forest and tree plans, including those outlined in the United Nations’ State of the World’s Forests technical report [1].
Despite its widespread use, mapping CC does not differentiate between individual tree crowns. This limitation is significant because CC mapping is unable to effectively convey other crucial characteristics such as variations in crown sizes, individual and collective tree locations, species, or the health of individual trees that are needed for tree inventorying and monitoring, which plays a crucial role in urban planning, environmental conservation, and forestry management.
Traditional methods for tree inventorying are time-consuming, labour-intensive, and often expensive. With the advent of remote sensing technologies and deep learning, it is now possible to leverage digital public orthoimages and artificial intelligence to automate and enhance the tree inventorying process [2] (Figure 1).
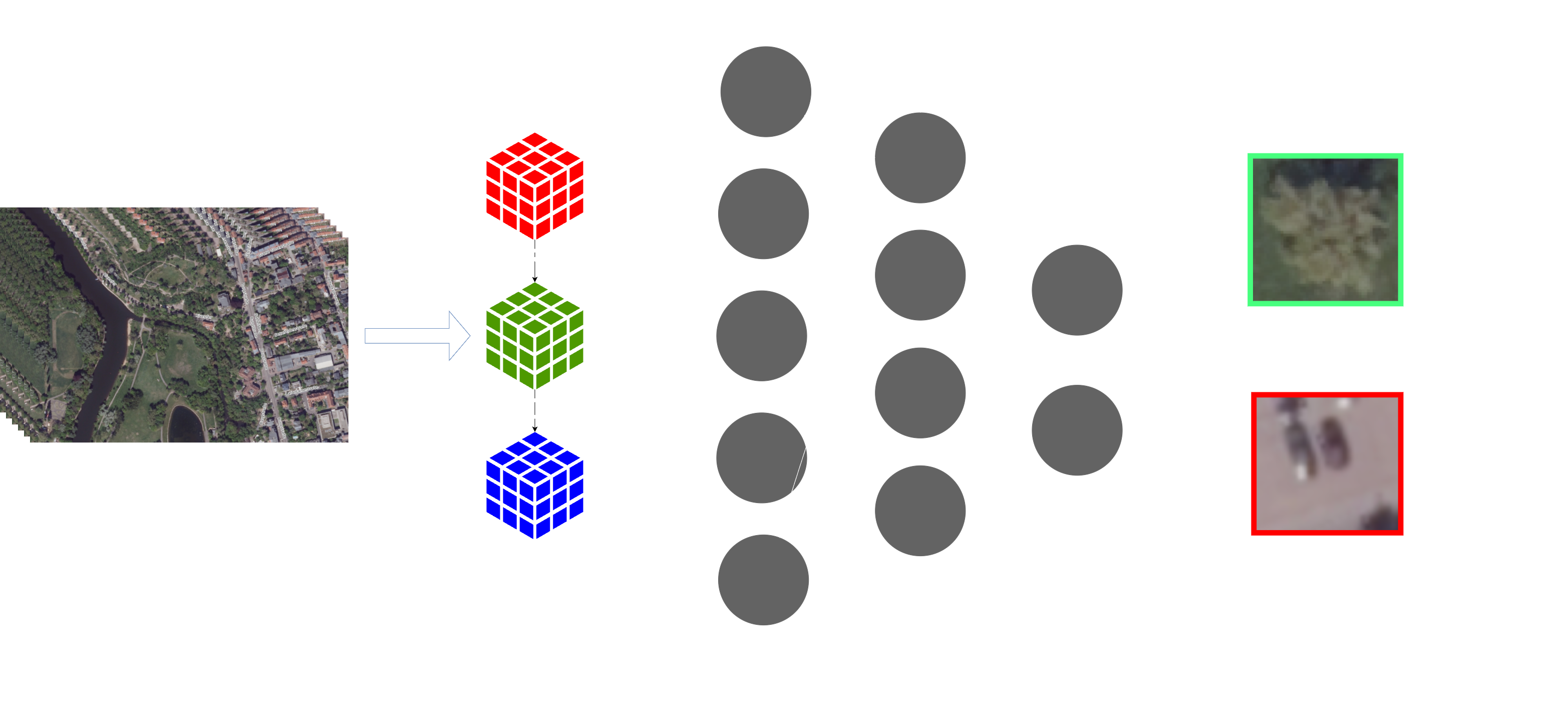
Segmented tree crowns can give a wealth of information about the individual trees (Figure 2). The significance of a tree’s fitness is closely tied to both the structure and size of its crown [3]. These factors influence the tree’s access to resources, its utilisation and occupation of space, the magnitude of its growth, as well as seed production and dispersal. In densely populated environments, the growth of crown size results in competition for space, leading to social differentiation, reduced growth of suppressed trees, mortality, and self-thinning [4]. Furthermore, the practical and economic relevance of crown structure and size should not be overlooked. Wider crowns contribute to heightened mechanical stability due to lower slenderness (h/d ratios), but this comes at the expense of wood quality, influenced by the number and thickness of branches [5].
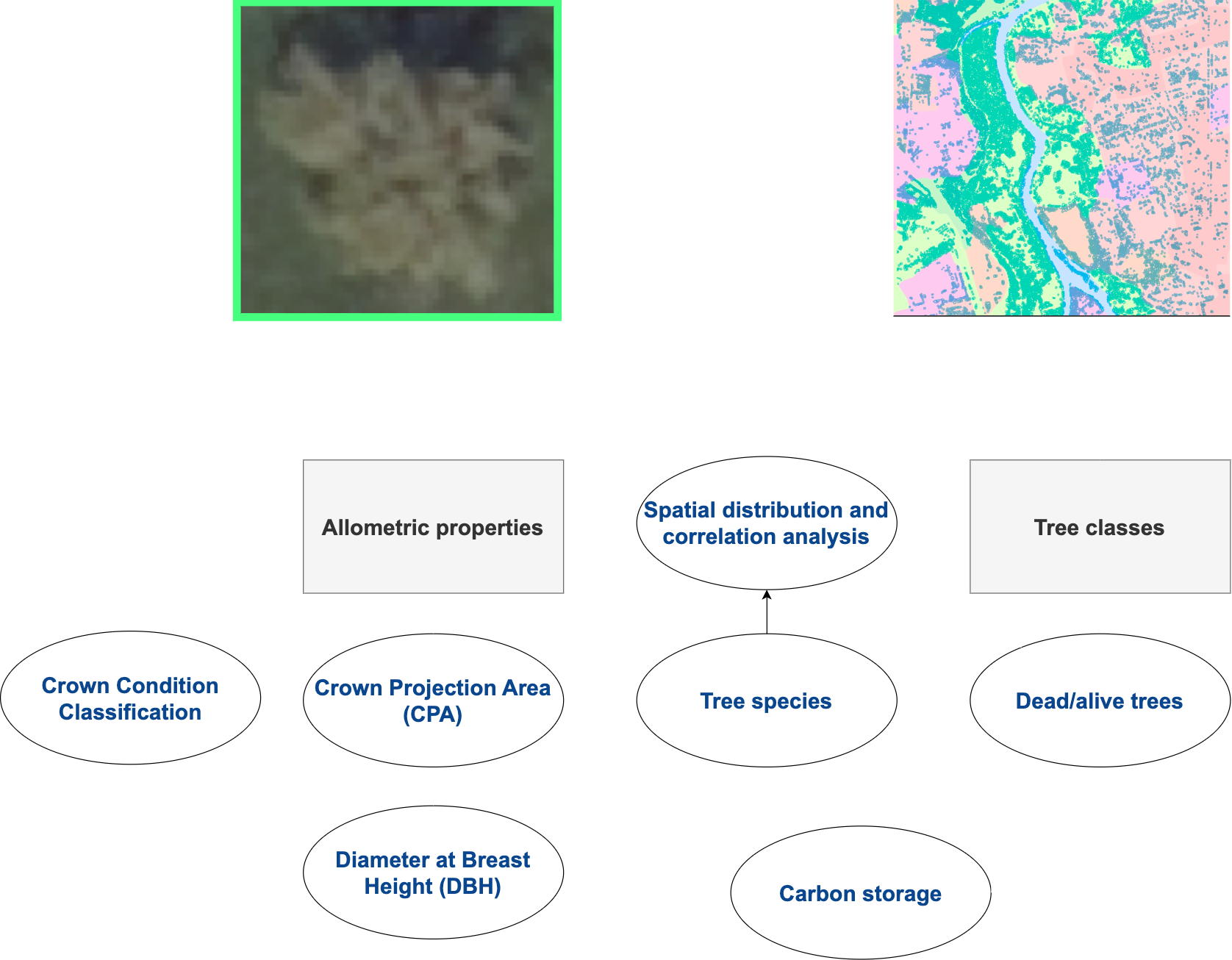
This finer level of detail holds importance in the analysis, modelling, or management of diverse environments, spanning natural forests, planted forests, urban forests, and orchards (Figure 2). Thus, mapping of individual tree crowns has proven to enhance the precision of carbon storage estimation [6], biodiversity assessment [7], urban forest management [8, 9], canopy closure estimation [10], ecosystem service modelling [11], and forest health description [12].
DeepTrees is a toolkit of workflows that standardise tree crown detection and analysis methods, and allows the workflows to be used to contribute to a centralised tree database (location independent). The project is geared towards citizens, researchers, urban planners, conservationists and activists who want to delineate tree crowns from ortho-/satellite imagery. DeepTrees will have three components major:
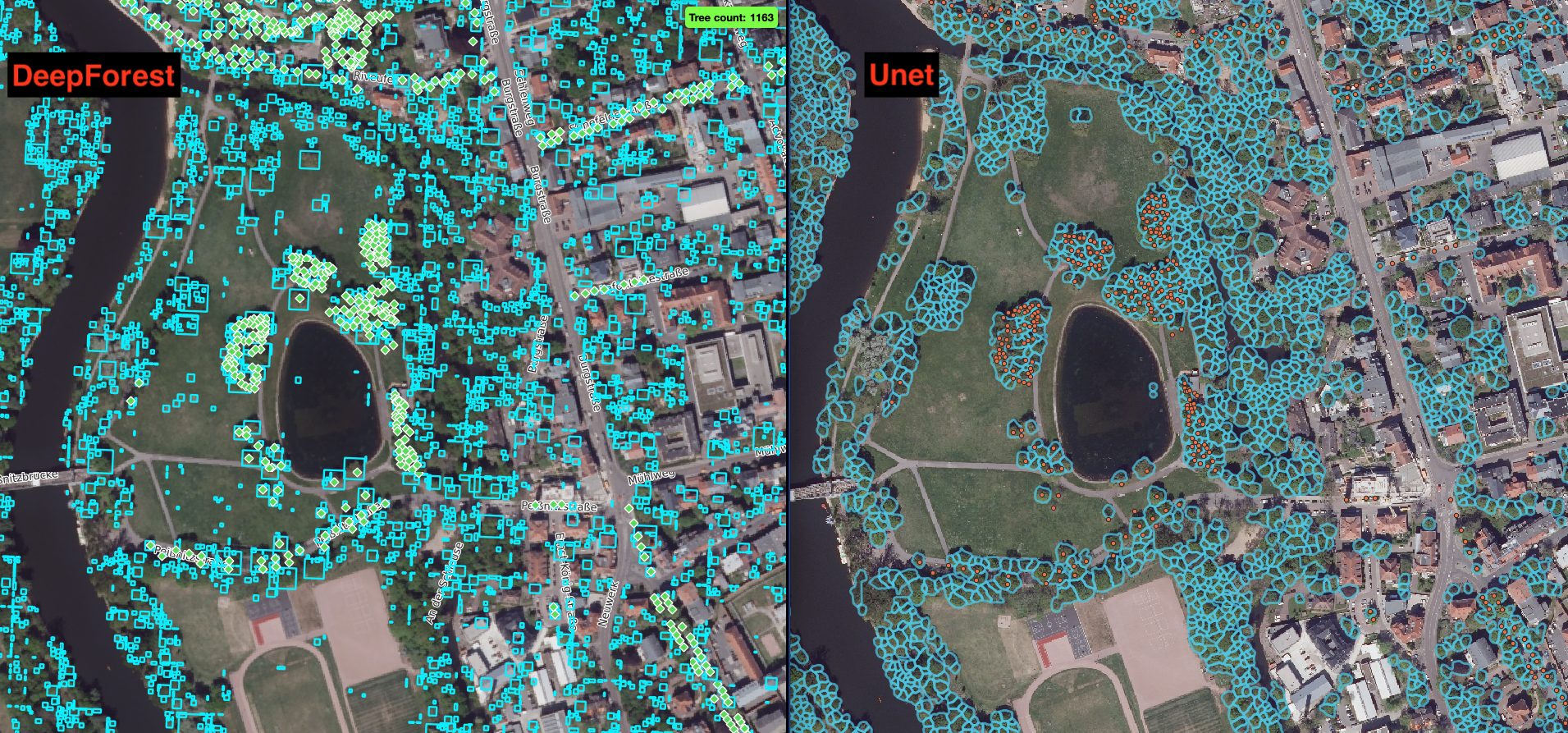
A set of publicly available workflows that allow users: to aggregate training and prediction of multiple deep learning models for standardised tree inventorying. to analyse the delineated trees (tree allometry + tree classes). A public database of trees where users can add modelled trees from their own workflow runs. An example tree monitoring system for the city of Halle (Saale) created using DeepTrees (Figure 3).
The DeepTrees project is a continuation of the Halle Treecrowns project, the results of which can be found on https://halletrees.de or https://haleetrees.de/about.
Digital Orthoimages Program (DOP)
The Digital Orthoimages Program (DOP) is a national program in Germany that aims to provide high-resolution orthoimages for the entire country. The DOP is a valuable data resource that can for large-scale tree inventorying and monitoring over time, as it offers detailed imagery that can be used to detect and delineate tree crowns. By leveraging the DOP data, DeepTrees aims to develop deep learning models that can accurately segment tree crowns and classify tree species, providing valuable information for forest management, biodiversity monitoring, and ecological research.
The focus of DeepTrees is on the 20 cm DOP imagery from the Federal States of Sachse-Anhalt and Sachse, although the workflows can be adapted to other regions and datasets.
Segmentation of Tree Crowns
These modeled tree crowns exhibit several distinctive characteristics that enable accurate identification and delineation of tree properties within imagery.

Firstly, the crowns mimic the intricate shapes of the tree crowns in the image. Through extensive training on diverse datasets, the models can learn to delineate the boundaries of tree crowns at a certain accuracy, distinguishing them from surrounding background elements. This capability is essential for applications such as forest inventory, where precise delineation of tree crowns is crucial for estimating biomass and species composition.
Texture analysis is another key aspect of tree crown segmentation. Tree crowns often exhibit distinct textural patterns, such as variations in foliage density and arrangement. The extracted polygons exhibit textural features, enabling differentiation between different parts of the crown and accurately segment them from non-crown elements in the image. Textures, combined with spectral signatures, can also enable classification of tree species based on their unique foliage patterns, providing valuable information for ecological studies and biodiversity monitoring (Figure 2.) [13].


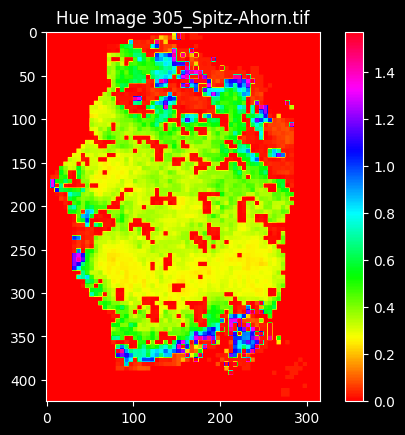
Furthermore, there are clear variations in color within tree crowns. Foliage color can vary significantly depending on factors such as species, health, and seasonal changes. From the segmented crown, the models can learn to recognize and segment these color variations, enhancing the accuracy of crown segmentation.
Edge detection is also critical for precise tree crown segmentation. The segmented crowns represent crown edges, even in complex and cluttered environments. This enables delineation of the boundary between tree crowns and their surroundings with high precision, facilitating applications such as forest mapping and ecological research.
Tree Traits
Leaf Chlorophyll Content (LCI)
We can estimate leaf chlorophyll content based on spectral reflectance data. By analyzing the spectral signatures of tree crowns, LCI can be used to infer to the chlorophyll content of leaves, providing valuable information about tree health and photosynthetic activity.

Crown Allometry
Tree crowns in many shapes and sizes. However, they can be summarized in 5 broad categories (Figure 3) [14].
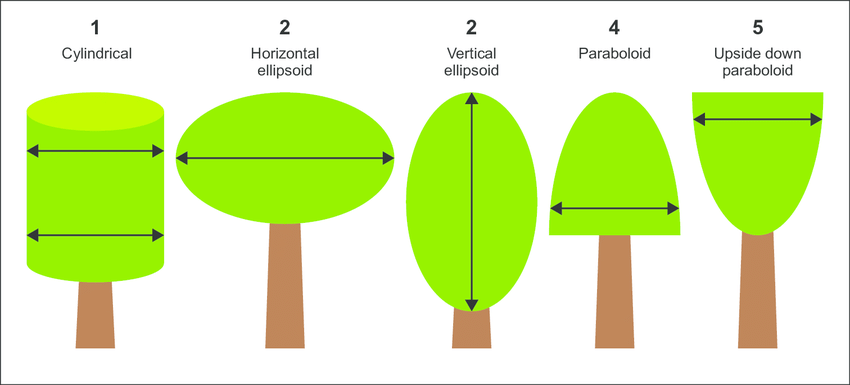
Segmented crowns can be used for analyzing tree crown allometry by using segmentation polygons of tree crowns and extracting relevant morphological features. These features carry inherent information about the size and shape of tree crowns across an area. By accurately delineating tree crowns from background elements, we can enable the extraction of more precise measurements related to crown size, such as crown diameter, area, and perimeter.
Segmentations can also extract features related to tree attributes, such as height and diameter, from remote sensing data or ground-based measurements. By integrating these attributes with crown morphology data, we can analyze the relationship between crown size, shape, and tree characteristics, contributing to the study of tree crown allometry.
Species Classification
The segmented crowns can also be used for species classification based on spectral signatures and textural features. By analyzing the unique spectral and textural characteristics of tree crowns, we can classify tree species. This capability is essential for applications such as biodiversity monitoring, forest management, and ecological research, where accurate species identification is crucial.
Phenological Analysis
The segmented tree crowns can provide valuable insights into tree phenology by analyzing changes in crown structure and color over time. By tracking the seasonal variations in tree crowns, we can identify phenological stages, such as leaf emergence, senescence, and flowering. This information is essential for studying the impact of climate change on tree growth and health.
The DOP data is available for both spring and summer months, which can be used to study phenological changes in tree crowns.
Applications
Forest Inventory
DeepTrees can be used for large-scale tree inventorying and monitoring in forests. By accurately segmenting tree crowns from orthoimages, DeepTrees can provide detailed information about tree species, size, and distribution. This information is essential for forest management, biomass estimation, and ecological research.
Crown Change
Additionally, segmentations in time series can be used to study adaptability of trees to various environmental conditions, including lighting, weather, and seasonal changes. Through robust training on diverse datasets, the underlying models can learn to adapt to changing environmental conditions, enhancing their generalization capabilities and ensuring reliable performance across different scenarios.
Urban Tree Management
Urban tree management is another key application of DeepTrees. By analyzing tree crowns in urban areas, DeepTrees can provide valuable information about tree health, species composition, and canopy cover. This information is essential for urban planners, conservationists, and policymakers to make informed decisions about tree planting, maintenance, and removal.
References
- FAO (2022). The State of the World’s Forests. Crossref, https://doi.org/10.4060/cb9360en.
- Zhao, H., Morgenroth, J., Pearse, G., & Schindler, J. (2023). A Systematic Review of Individual Tree Crown Detection and Delineation with Convolutional Neural Networks (CNN). Current Forestry Reports, 1-22.
- White, E. P., Ernest, S. M., Kerkhoff, A. J., & Enquist, B. J. (2007). Relationships between body size and abundance in ecology. Trends in ecology & evolution, 22(6), 323-330.
- Knoke, T., Ammer, C., Stimm, B., & Mosandl, R. (2008). Admixing broadleaved to coniferous tree species: a review on yield, ecological stability and economics. European journal of forest research, 127, 89-101.
- Pretzsch, H., & Rais, A. (2016). Wood quality in complex forests versus even-aged monocultures: review and perspectives. Wood science and technology, 50, 845-880.
- Fujimoto, A., Haga, C., Matsui, T., Machimura, T., Hayashi, K., Sugita, S., & Takagi, H. (2019). An end to end process development for UAV-SfM based forest monitoring: Individual tree detection, species classification and carbon dynamics simulation. Forests, 10(8), 680.
- Saarinen, N., Vastaranta, M., Näsi, R., Rosnell, T., Hakala, T., Honkavaara, E., … & Hyyppä, J. (2018). Assessing biodiversity in boreal forests with UAV-based photogrammetric point clouds and hyperspectral imaging. Remote Sensing, 10(2), 338.
- Kimball, L. L., Wiseman, P. E., Day, S. D., & Munsell, J. F. (2014). Use of urban tree canopy assessments by localities in the Chesapeake Bay Watershed. Cities and the Environment (CATE), 7(2), 9.
- Berland, A., Shiflett, S. A., Shuster, W. D., Garmestani, A. S., Goddard, H. C., Herrmann, D. L., & Hopton, M. E. (2017). The role of trees in urban stormwater management. Landscape and urban planning, 162, 167-177.
- Brūmelis, G., Dauškane, I., Elferts, D., Strode, L., Krama, T., & Krams, I. (2020). Estimates of tree canopy closure and basal area as proxies for tree crown volume at a stand scale. Forests, 11(11), 1180.
- Livesley, S. J., McPherson, E. G., & Calfapietra, C. (2016). The urban forest and ecosystem services: Impacts on urban water, heat, and pollution cycles at the tree, street, and city scale. Journal of environmental quality, 45(1), 119-124.
- Shendryk, I., Broich, M., Tulbure, M. G., McGrath, A., Keith, D., & Alexandrov, S. V. (2016). Mapping individual tree health using full-waveform airborne laser scans and imaging spectroscopy: A case study for a floodplain eucalypt forest. Remote Sensing of Environment, 187, 202-217.
- Maschler, J., Atzberger, C., & Immitzer, M. (2018). Individual tree crown segmentation and classification of 13 tree species using airborne hyperspectral data. Remote Sensing, 10(8), 1218.
- McPherson, E. G., van Doorn, N. S., & Peper, P. J. (2016). Urban tree database and allometric equations (Vol. 253). Albany, CA, USA: US Department of Agriculture, Forest Service, Pacific Southwest Research Station.